March 1, 2017
3 Bullets:
- Identifying driver mutations in cancer has been a major challenge in cancer research, with the ultimate goal of understanding the detailed molecular origins of cancer and providing genetically personalized treatments.
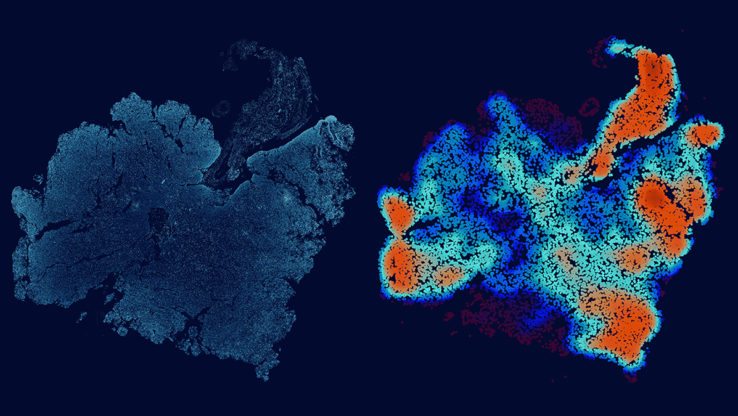
- This novel clustering algorithm bridges the gap between a focus on single amino acid mutations and mutations observed anywhere in the entire gene by identifying variable length regions within cancer genes which are enriched for mutations.
- Integration of the multiscale mutation clusters with gene expression data will help to gain insight into the functional consequences of these mutations.
By Theo Knijnenburg
The identification of mutations that alter the function of protein-coding genes in cancer and a molecular understanding of the ensuing consequences of such mutations remains a significant challenge. To date, millions of distinct somatic mutations have been observed in human cancers through genome wide characterization projects such as The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC). Computational methods are particularly well-suited for the assessment of somatic mutations at this scale in order to identify those with cancer-associated functional consequences.
In a study published in PLoS Computational Biology, researchers at Institute for Systems Biology (ISB) have developed a multiscale mutation clustering algorithm (M2C) that identifies variable length regions with high mutation density in cancer genes. They applied their algorithm on hundreds of frequently mutated genes using the combined mutation data in over twenty tumor types from TCGA and identified over a thousand multiscale mutation clusters. Statistical association of these multiscale clusters with gene expression from TCGA tumor samples and drug response data from cancer cell lines, illuminates the (differential) functional and associated therapeutic consequences of somatic mutations in cancer.
The M2C algorithm was developed by William Poole (first author on the paper), who started as a summer intern in 2013 as part of ISB’s Center for Systems Biology internship program. Working under the guidance of Dr. Brady Bernard and Dr. Theo Knijnenburg, both senior research scientists in the lab of Ilya Shmulevich at ISB, Poole’s initial summer internship led to a multi-year project that resulted not only in this PLoS Computational Biology publication, but also a Bioinformatics publication about combining dependent P-values. His work was presented at two international scientific conferences: the TCGA Scientific Symposium 2015 and The 15th European Conference on Computational Biology (ECCB 2016). Currently, Poole is pursuing a PhD at the California Institute of Technology (Caltech) in Pasadena, California.
Title: Multiscale mutation clustering algorithm identifies pan-cancer mutational clusters associated with pathway-level changes in gene expression
Journal: PLOS Computational Biology
Authors: William Poole, Kalle Leinonen, Ilya Shmulevich, Theo A. Knijnenburg, Brady Bernard
Link: journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005347


 isbscience.org/research/novel-clustering-algorithm-identifies-functional-mutations-cancer-genes/
isbscience.org/research/novel-clustering-algorithm-identifies-functional-mutations-cancer-genes/