What is Systems Biology
Systems biology is based on the understanding that the whole is greater than the sum of the parts.
Systems biology has been responsible for some of the most important developments in the science of human health and environmental sustainability. It is a holistic approach to deciphering the complexity of biological systems that starts from the understanding that the networks that form the whole of living organisms are more than the sum of their parts. It is collaborative, integrating many scientific disciplines – biology, computer science, engineering, bioinformatics, physics and others – to predict how these systems change over time and under varying conditions, and to develop solutions to the world’s most pressing health and environmental issues.
This ability to design predictive, multiscale models enables our scientists to discover new biomarkers for disease, stratify patients based on unique genetic profiles, and target drugs and other treatments. Systems biology, ultimately, creates the potential for entirely new kinds of exploration, and drives constant innovation in biology-based technology and computation.
Because systems biology requires constant attention to a very complex, very human social experiment, ISB fosters the kind of financial, social and psychological environment in which the world’s best scientists, technologists, engineers and mathematicians can collaborate and do their best work.
Dr. Nitin Baliga, ISB SVP and Director, explains what is systems biology. Watch on YouTube.
SYSTEMS BIOLOGY 101:
ISB’s Innovation Engine
A fundamental tenet of systems biology is that solving challenging biological problems always requires the development of new technologies in order to explore new dimensions of data space. New data types require novel analytical tools. This virtuous cycle of biology driving technology driving computation can exist only in a cross-disciplinary environment where biologists, chemists, computer scientists, engineers, mathematicians, physicists, physicians and others can come together in teams to tackle grand challenges. This is ISB. And this describes what we call the “innovation engine” (depicted below) that drives our ability to develop intellectual property, which we share through open-access platforms or by spinning out companies.
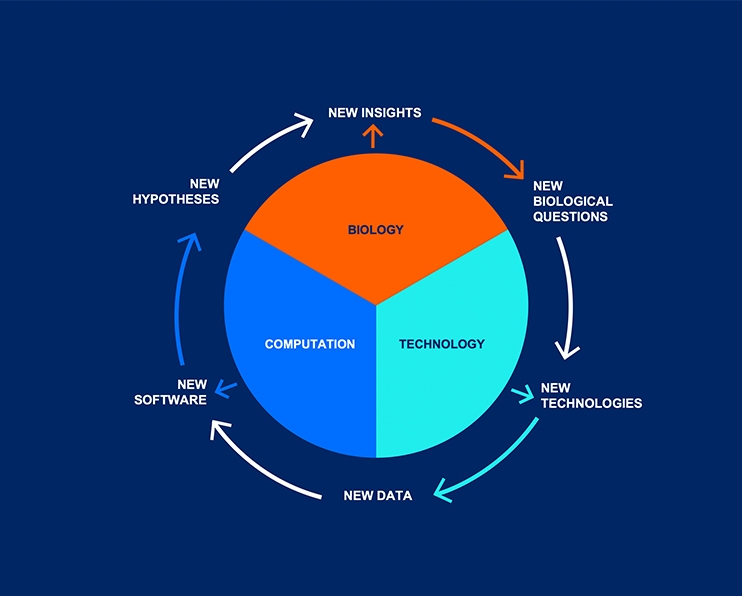
Cross-disciplinary Teams
In describing systems biology and the distinguishing characteristics of ISB’s approach, we always emphasize how our lab groups are intentionally and necessarily cross-disciplinary. One of our labs, for example, includes molecular biologists, microbiologists, geneticists, engineers, oceanographers, and even an astrophysicist. The complexity of biology in this age of “big data” requires diverse teams in order to tackle such vast amounts of data and to make sense of it all. New technologies that crunch data faster and more efficiently also permit researchers to re-analyze existing datasets, a process which often reveals undiscovered information. Complementary skills empower any of our groups of researchers to better understand biological or environmental challenges from different perspectives and to arrive at shareable insights more quickly. Our interdisciplinary teams have contributed notable advances to everything from ocean acidification to neurodegenerative diseases and tuberculosis to multiple cancers.
Network of Networks
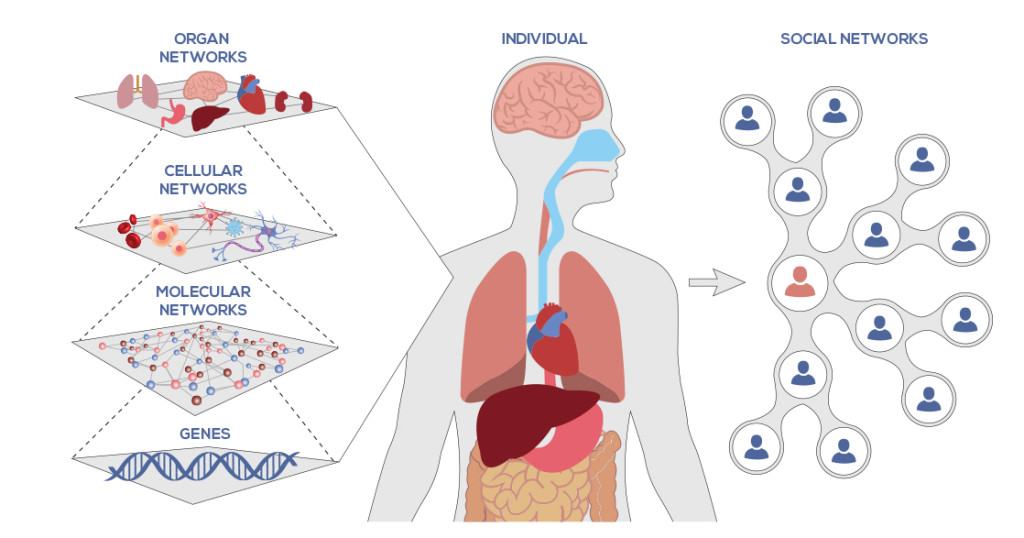 With networks, we can organize and integrate information at different levels. Social networks have transformed communications in the 21st Century, democratizing our platforms for communications. At ISB we are also concerned with networks. One of the tenets of systems biology we often refer to is the “Network of Networks.” On a biological level, our bodies are made up of many networks that are integrated at and communicating on multiple scales. From our genome to the molecules and cells that makeup the organs in our bodies all the way out to ourselves in our world: we are fundamentally a network of networks. Systems biology looks at these networks across scales to integrate behaviors at different levels, to formulate hypotheses for biological function and to provide spatial and temporal insights into dynamical biological changes. It is not enough to understand only one part of a system when studying the complexity of biology. Therefore the framework of the “Network of Networks” provides meaningful insight into understanding how systems biology’s approach is different, more integrated and more capable of analyzing and predicting state transitions in biological systems.
With networks, we can organize and integrate information at different levels. Social networks have transformed communications in the 21st Century, democratizing our platforms for communications. At ISB we are also concerned with networks. One of the tenets of systems biology we often refer to is the “Network of Networks.” On a biological level, our bodies are made up of many networks that are integrated at and communicating on multiple scales. From our genome to the molecules and cells that makeup the organs in our bodies all the way out to ourselves in our world: we are fundamentally a network of networks. Systems biology looks at these networks across scales to integrate behaviors at different levels, to formulate hypotheses for biological function and to provide spatial and temporal insights into dynamical biological changes. It is not enough to understand only one part of a system when studying the complexity of biology. Therefore the framework of the “Network of Networks” provides meaningful insight into understanding how systems biology’s approach is different, more integrated and more capable of analyzing and predicting state transitions in biological systems.
Multiscale Modeling
Whether we explicitly recognize it or not, multiscale phenomena are part of our daily lives. We organize our time in days, months and years, as a result of the multiscale dynamics of the solar system. Our society is organized in a hierarchical structure, from towns to states, countries and continents. The human body is a complex machine, with many little parts that work by themselves or with other parts to perform specific functions. Organelles inside each cell in our bodies interact with one another to maintain a healthy functioning cell that moves, differentiates and dies. These subcellular organelles and their processes govern each cell’s signalling mechanism to interact with its neighboring cells, and form multi-cellular systems called tissues (e.g. epithelial tissue, muscular tissue). Two or more types of tissues work together to form an organ that performs a specific task (e.g. mouth, stomach, liver). Two or more organs work together to form organ systems, such as the digestive system and the nervous system, that perform more complex tasks. All these organ systems interact with each other to enable a healthy functioning organism. Traditional approaches to modeling real world systems focus on a single scale that imparts a limited understanding of the system. The pace at which biotechnology has grown has enabled us to collect large volumes of data capturing behavior at multiple scales of a biological system. Genetic as well as environmental alterations to the DNA, expression levels of RNAs, expression of genes and synthesis of proteins – all this is measurable now within a matter of days at a rapidly declining cost. So, it is really up to scientists and data analysts to make use of this variety of data types and build integrative models that enable a comprehensive understanding of the system under study. Multiscale models do just that. By integrating models at different scales and allowing flow of information between them, multiscale models describe a system in its entirety, and as such, are intrinsic to the principles of systems biology.
Single Cell Analysis
It is well known that there is no “average” patient. Therefore, in clinical trials encompassing large groups of patients, one needs to consider the characteristics of each patient, including each person’s individual genetic propensity to respond to a drug in a particular way. The statistical analysis of population averages suppresses valuable individual-specific information. The consideration of population heterogeneity due to inevitable patient-to-patient variability is called “stratification” and is at the heart of personalized medicine. Such stratification will allow a proper impedance match against appropriate and effective drugs. Each cell in a cell population of apparently identical cells is a distinct individual. There is no “average” cell even within a population of cells of the very same cell type. Just as one can look at individual patients in a population and identify subtypes of diseases, one can identify “quantized” or “discrete” cell-subtypes in a cell population. The quantized subtypes perform different functions and form a network – much like a social network in human populations. So understanding how an organ works will require understanding the coordinated integration of the functioning of all the quantized cell types. Because of such cellular heterogeneity, even the most potent target-selective drug will kill only a fraction of tumor cells – explaining the inexorable drug resistance in malignant tumors. This new insight on cellular heterogeneity calls for the measurement of all molecular profiles in individual cells. Tissues must be seen not as an amorphous mass but analyzed as dynamical populations of cells and at single-cell resolution.
Understanding Proteomics
If DNA is the blueprint for life, then proteins are the bricks. The genes in DNA are translated into proteins, strings of amino acids that fold into three-dimensional structures. The type and order of the amino acids in a protein will change its shape and determine its special function. Proteins are the molecules that make life happen: they are the powerplants that turn food into energy, the machines that make cells move, and even the computers that read DNA and make more proteins. The information to build every protein in an organism is contained in the DNA, but not every protein is produced at once or in the same amount. Think of a cell in your liver and a cell in your retina – both cells contain identical DNA, but very different subsets of proteins are being produced in order to give each cell its special function. Proteomics is the discipline of identifying and quantifying the proteins present in an organism. At ISB, we use state-of-the-art scientific instruments and cutting edge computational techniques to detect thousands of proteins at once, giving us a systems-level view of the molecular machinery of life.
Be at the forefront of profound breakthroughs in human health. Be a part of ISB and the revolution we’re proud to have helped cause.
Our progress requires you.



